New Research Identifies Four Unique Autism Subtypes with Genetic Links
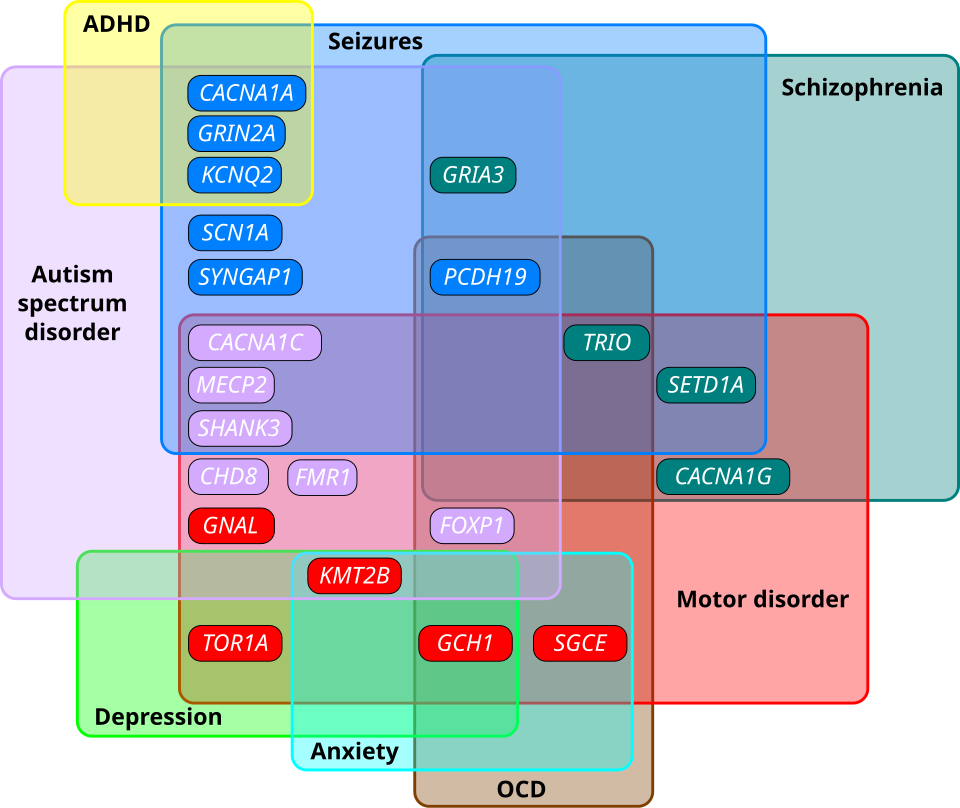
A groundbreaking study published in Nature Genetics has identified four distinct subtypes of autism, each characterized by unique genetic signatures and phenotypic traits. This research, led by scientists at the Flatiron Institute's Center for Computational Biology (CCB) and collaborators, utilized data from SPARK, the largest autism study to date, encompassing over 5,000 participants aged 4-18. The findings, which emerged from the collaboration of multiple prominent institutions including Princeton University and the Simons Foundation, aim to enhance support for individuals with autism by linking observable traits to underlying genetics.
Understanding autism as a spectrum is essential due to its diverse manifestations, which can complicate diagnosis and treatment. According to Natalie Sauerwald, a CCB associate research scientist and co-lead author of the study, "A clinically grounded, data-driven subtyping of autism would really help kids get the support they need early on". The study’s results suggest that recognizing these subtypes can enable caregivers to access tailored resources and interventions, particularly for associated conditions such as ADHD and anxiety.
The research identified four main groups based on shared traits: 1. **Social and Behavioral Challenges** (37% of participants): Individuals in this group exhibit co-occurring conditions like ADHD, anxiety disorders, and mood dysregulation, yet display typical developmental milestones. 2. **Mixed ASD with Developmental Delay** (19%): This group shows developmental delays but lacks significant anxiety or behavioral issues. 3. **Moderate Challenges** (34%): Participants experience challenges similar to the first group but to a lesser extent and without developmental delays. 4. **Broadly Affected** (10%): This group faces widespread challenges, including severe communication difficulties and mood disorders.
The researchers utilized advanced data analysis techniques, including general finite mixture modeling, to categorize participants effectively while focusing on a person-centered approach. This methodology contrasts with traditional trait-centered studies, allowing for a more holistic understanding of autism.
"Our model allowed us to define groups of individuals with shared phenotypic profiles, which translates to clinically similar presentations," stated Aviya Litman, another lead author of the study.
The genetic analysis revealed that each subtype is influenced by different genetic pathways, which affect biological processes in distinct ways. For instance, the Social and Behavioral Challenges group had genetic variants primarily active after birth, while those in the Mixed ASD with Developmental Delay group showed significant prenatal gene activity.
Kelsey Martin, executive vice president of autism and neuroscience at the Simons Foundation, emphasized the significance of large datasets in making these discoveries: "This study underscores the power of leveraging machine learning approaches to analyze the vast amount of data available in SPARK."
The research team hopes that their findings will pave the way for future studies that delve deeper into the non-coding regions of the genome, which are known to play critical roles in gene regulation but remain less explored. "The more data, the more discovery," said Sauerwald, highlighting the potential for further insights into autism's genetic underpinnings.
This study is not just a scientific milestone; it represents a crucial step towards developing personalized interventions tailored to the unique profiles of individuals on the autism spectrum. As the understanding of autism continues to evolve, these findings may significantly improve the quality of life for many affected individuals and their families.
Advertisement
Tags
Advertisement