Scientists Develop New Method to Enhance Fluorescence Microscopy Resolution
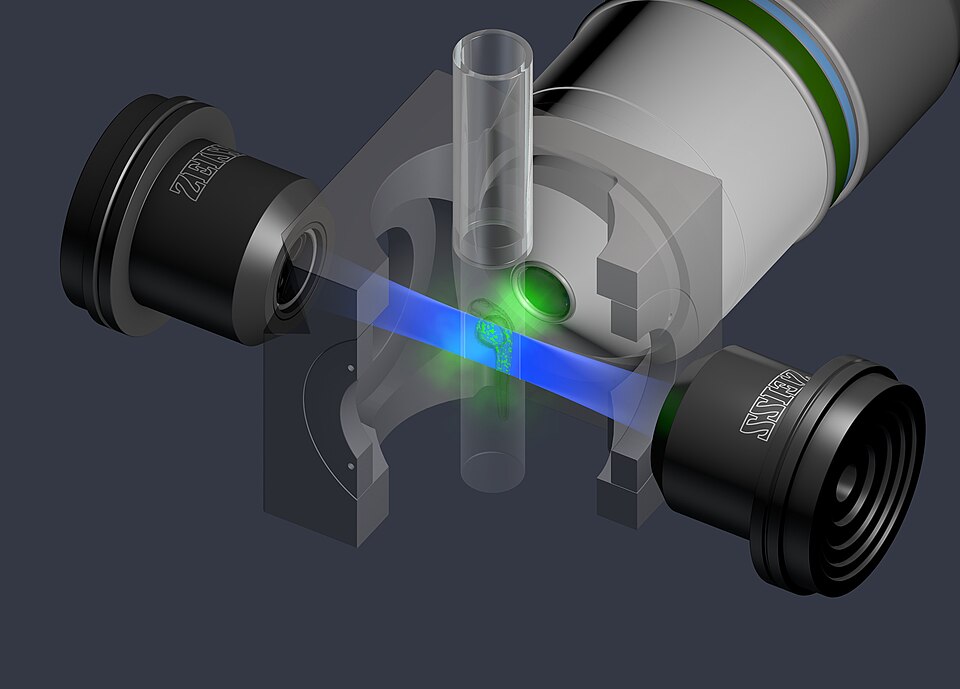
In a groundbreaking achievement, researchers from the Janelia Research Campus have unveiled a new method for fluorescence microscopy that significantly enhances its resolution capabilities. This advancement, reported on June 17, 2025, aims to improve the accuracy of molecular counts in biological studies, potentially transforming the landscape of cellular biology and biochemistry.
Fluorescence microscopy, a widely used technique in biological research, allows scientists to visualize and study dynamic processes in living cells. Traditionally, the resolution of fluorescence microscopes is limited by optical diffraction, which prevents researchers from distinguishing closely spaced molecules. The new technique, developed by the Funke Lab at Janelia, addresses this limitation by utilizing advanced computational modeling and machine learning.
According to Jan Funke, Group Leader at Janelia and one of the lead researchers, "Our method, named 'blinx,' enables us to infer the number of individual molecules within a single spot of light captured by the microscope. Just as one might listen to the varied croaks of frogs to determine their numbers, our approach analyzes fluctuations in fluorescence intensity to estimate molecular counts."
The core innovation behind blinx involves modeling the entire light path through the microscopy system—from the moment photons exit the fluorophore to their detection in the microscope. By generating a trace of light intensity over time, researchers can adjust model parameters to match experimental data, ultimately revealing the probability distribution of molecule counts rather than a single, uncertain estimate.
Dr. Sarah Johnson, Professor of Biology at Stanford University, emphasizes the significance of this development: "The ability to provide probability distributions enhances our understanding of molecular dynamics in cells. This could be especially critical in identifying proteins and their interactions in complex biological systems."
The implications of blinx are vast. Previous methods often yielded ambiguous results, lacking the robustness necessary for confident biological interpretation. By contrast, blinx not only improves counting accuracy but also allows researchers to gauge the reliability of their data. Funke elaborates, "This method can indicate when the data is insufficient to yield a clear answer, which is crucial for guiding further investigation."
Moreover, as Alex Hillsley, a former postdoctoral researcher in the Funke and Stern labs, notes, the interdisciplinary nature of the project was instrumental to its success. "Developing blinx required expertise in chemistry, super-resolution imaging, and computational modeling—skills that coalesce uniquely at Janelia."
The potential applications of blinx extend beyond mere molecular counting. Researchers anticipate its utility in areas such as drug development, where precise protein quantification could lead to more effective therapeutic strategies. As global health challenges continue to evolve, advancements like these in microscopy techniques represent critical steps toward understanding complex biological systems.
Looking ahead, Funke expressed hope that blinx will stimulate further innovations in microscopy methodologies. "This is just the beginning. I envision a new generation of algorithms that will build upon our findings, enhancing our ability to explore the microscopic world."
The research conducted at Janelia underscores the importance of collaborative environments in scientific discovery, where diverse expertise converges to tackle ambitious projects. As microscopy technology evolves, the potential for transformative discoveries in biology remains vast, promising to illuminate the intricate workings of life at the molecular level.
This research was supported by the Howard Hughes Medical Institute and highlights the ongoing commitment to advancing scientific knowledge through innovative technology and collaborative research efforts.
Advertisement
Tags
Advertisement





